Nusinersen
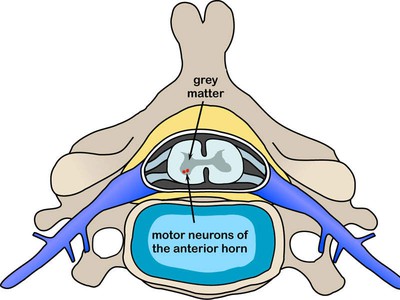
SMA(spinal muscular atrophy)는 spinal cord의 motor neuron의 loss로 인해 생기는 병이다.
Intron retention은 plant와 budding yeast에서는 gene expression regulation의 major mechanism으로 알려져 있었다. 최근까지도 mammalian system에서는 intron retention의 functional significance에 대해서는 의문점들이 있었다. 실험실에서 intron retention이 된 transcript를 찾는 등의 증거물을 확보하는 일은 기술적으로 어려운 일이었고, noise 정도의 수준으로 생각되는 정도였다. High-throughput deep sequencing 등의 기술의 발달도 정보가 넘쳐흐르자, physiological / pathological context에서 intron retention의 패턴에 대해 더 상세하게 분석할 수 있게 되었다.
정상적인 발달은 물론이고 스트레스나 질병에 대한 생체의 대응방식의 중요한 구성요소임이 최근 연구에서 밝혀졌다. 또한 intron retention이 어떠한 메커니즘으로 다음을 조절하는지 연구가 되고 있다.
- protein isoform production
- RNA stability and translation efficiency
- rapid induction of expression via post-transcriptional splicing of retained introns.
Both survival of motor neuron (*SMN*) genes are associated with spinal muscular atrophy; mutations in *SMN1* cause the disease, and *SMN2* modulates its severity
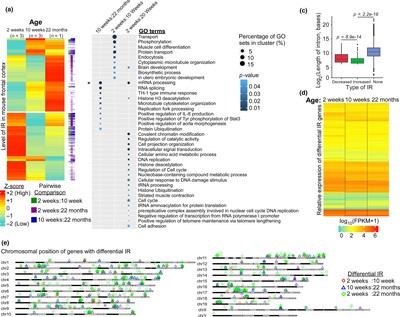
people are studying intron retention in mice brains, and the results are as follows:
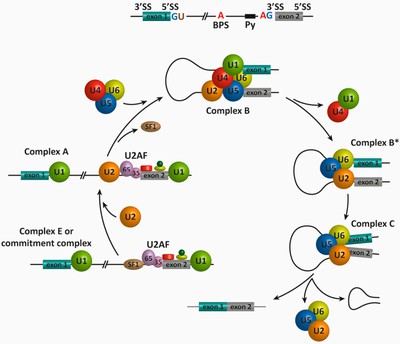
Splicing을 target하는 방법은 간단하다. 다른 diagram을 보자:
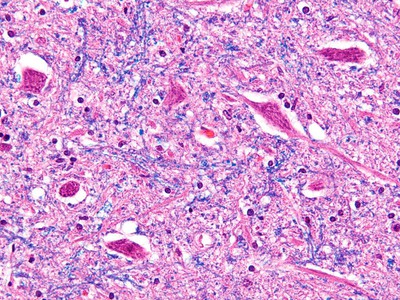
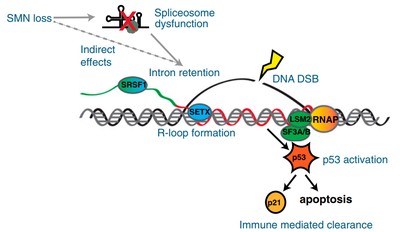
SMN deficiency는 SMA(spinal muscular atrophy)의 severe model에서 widespread한 intron retention과 DNA damage를 일으킨다고 한다.
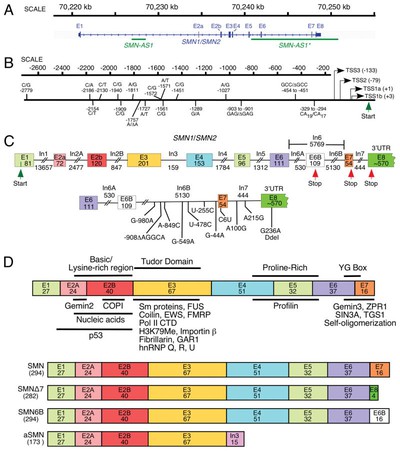
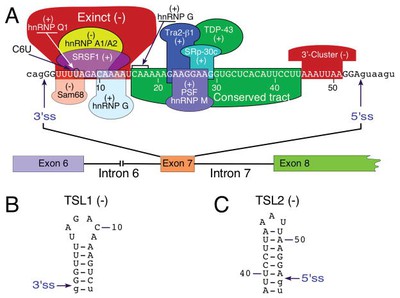
This is really cool.
References
Mohini Jangi et al., “SMN deficiency in severe models of spinal muscular atrophy causes widespread intron retention and DNA damage”, PNAS March 21, 2017 114 (12) E2347-E2356; first published March 7, 2017 https://doi.org/10.1073/pnas.1613181114
Jacob, A.G. & Smith, C.W.J. “Intron retention as a component of regulated gene expression programs”, Hum Genet (2017) 136: 1043. https://doi.org/10.1007/s00439-017-1791-x
Satomi Yoshimoto et al., “Alternative splicing of a cryptic exon embedded in intron 6 of SMN1 and SMN2”, Hum Genet Var (2016) 3: 16040. https://doi.org/10.1038/hgv.2016.40
Swarnaseetha Adusumalli et al., “Increased intron retention is a post‐transcriptional signature associated with progressive aging and Alzheimer’s disease”, IFAA, First published: 13 March 2019 https://doi.org/10.1111/acel.12928
Marc Suñé-Pou et al., “Targeting Splicing in the Treatment of Human Disease”, Genes 2017, 8, 87; https://doi.org/10.3390/genes8030087
Wurster CD, Ludolph AC. Nusinersen for spinal muscular atrophy. Ther Adv Neurol Disord. 2018;11:1756285618754459. Published 2018 Mar 13. doi:10.1177/1756285618754459
Other Links:
https://www.drugdevelopment-technology.com/projects/spinraza-nusinersen-for-the-treatment-of-spinal-muscular-atrophy-sma/